Wishbone
haraldschilly • • r
Do you want to run Wishbone in your SMC project?
Wishbone is an algorithm to align single cells from differentiation systems with bifurcating branches. Wishbone has been designed to work with multidimensional single cell data from diverse technologies such as Mass cytometry and single cell RNA-seq.
First, open a terminal file and run the following lines to install it locally inside your project:
pip3 install --user Cython
pip3 install --user git+https://github.com/dominiek/python-bhtsne.git
pip3 install --user git+https://github.com/jacoblevine/phenograph.git
pip3 install --user git+https://github.com/ManuSetty/wishbone.git
Then, open a new Jupyter notebook, switch to the “Python 3” kernel, and run examples from their documentation.
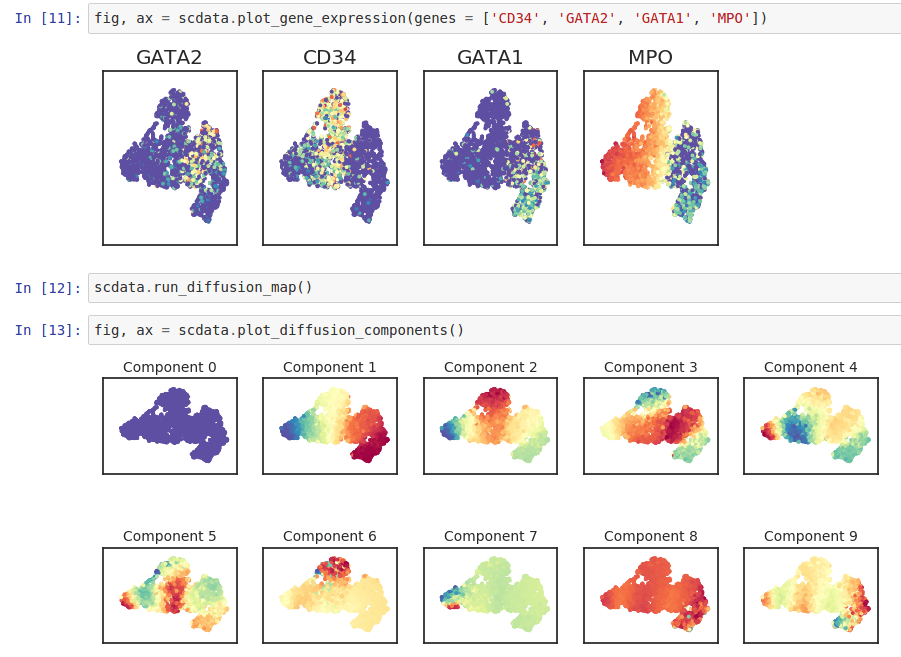
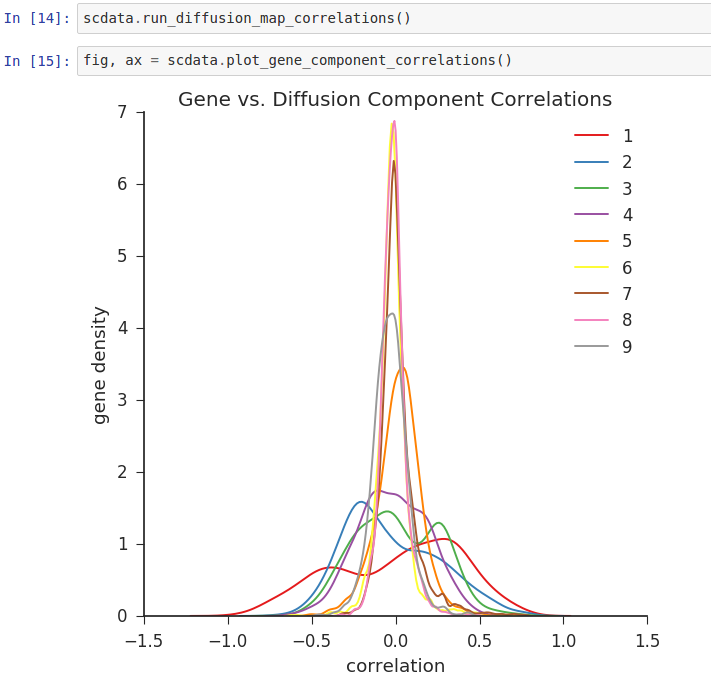
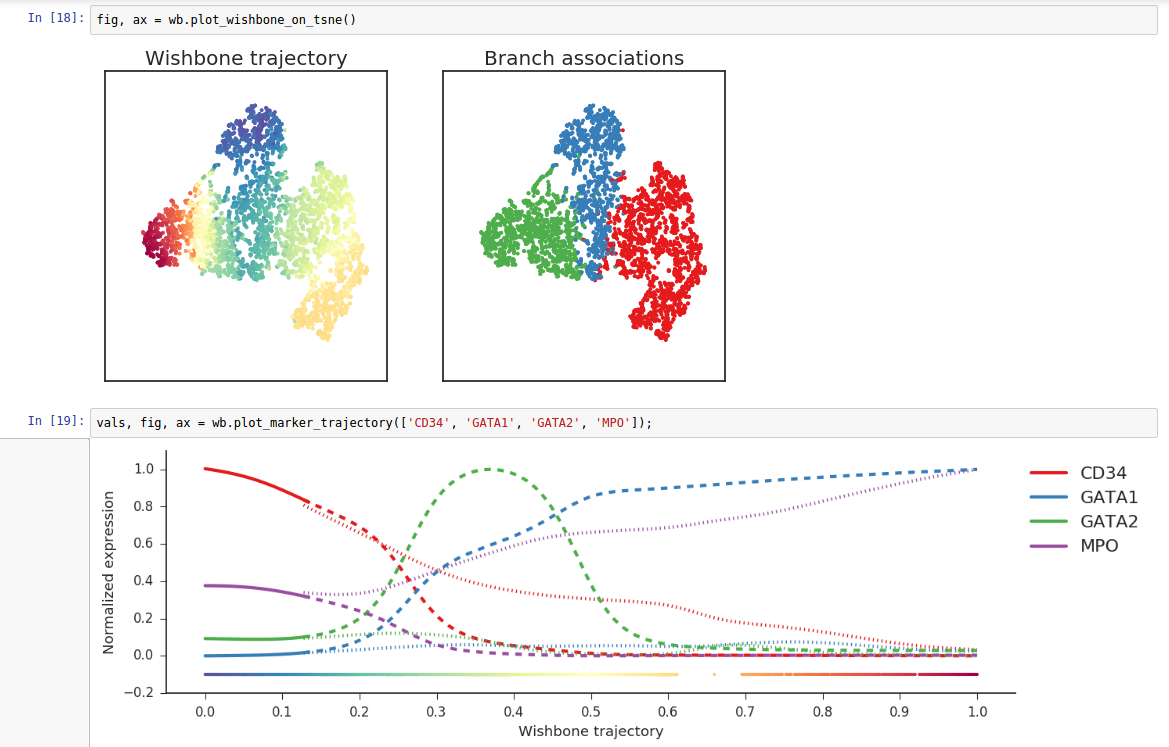
(In order to access GitHub or PyPi, your project needs “internet access”)